Knowing both the Field of View (FoV) of a camera’s lens and the dimensions of the object we’d like to measure (Region of Interest, ROI) seems like more than enough to get a distance.
Note, opencv has an extensive suite of actual calibration tools and utilities here.
…But without calibration or much forethought, could rough measurements of known objects even be usable? Some notes from a math challenged individual:
# clone:
git clone https://github.com/Jesssullivan/misc-roi-distance-notes && cd misc-roi-distance-notes
Most webcams don’t really provide a Field of View much greater than ~50 degrees- this is the value of a MacBook Pro’s webcam for instance. Here’s the plan to get a Focal Length value from Field of View:
&space;tan(\frac{FieldOfView}{2}))
So, thinking along the lines of similar triangles:
- Camera angle
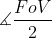 forms the angle between the hypotenuse side (one edge of the FoV angle) and the adjacent side
forms the angle between the hypotenuse side (one edge of the FoV angle) and the adjacent side
- Dimension
 is the opposite side of the triangle we are using to measure with.
is the opposite side of the triangle we are using to measure with.
- ^ This makes up the first of two "similar triangles"
- Then, we start measuring: First, calculate the opposite ROI Dimension using the arbitrary Focal Length value we calculated from the first triangle- then, plug in the Actual ROI Dimensions.
- Now the adjacent side of this ROI triangle should hopefully be length, in the the units of ROI’s Actual Dimension.
source a fresh venv to fiddle from:
# venv:
python3 -m venv distance_venv
source distance_venv/bin/activate
# depends are imutils & opencv-contrib-python:
pip3 install -r requirements.txt
The opencv people provide a bunch of prebuilt Haar cascade models, so let’s just snag one of them to experiment. Here’s one to detect human faces, we’ve all got one of those:
mkdir haar
wget https://raw.githubusercontent.com/opencv/opencv/master/data/haarcascades/haarcascade_frontalface_alt2.xml -O ./haar/haarcascade_frontalface_alt2.xml
Of course, an actual thing with fixed dimensions would be better, like a stop sign!
Let’s try to calculate the distance as the difference between an actual dimension of the object with a detected dimension- here’s the plan:

YMMV, but YOLO:
# `python3 measure.py`
import math
from cv2 import cv2
DFOV_DEGREES = 50 # such as average laptop webcam horizontal field of view
KNOWN_ROI_MM = 240 # say, height of a human head
# image source:
cap = cv2.VideoCapture(0)
# detector:
cascade = cv2.CascadeClassifier('./haar/haarcascade_frontalface_alt2.xml')
while True:
# Capture & resize a single image:
_, image = cap.read()
image = cv2.resize(image, (0, 0), fx=.7, fy=0.7, interpolation=cv2.INTER_NEAREST)
# Convert to greyscale while processing:
gray_conv = cv2.cvtColor(image, cv2.COLOR_BGR2GRAY)
gray = cv2.GaussianBlur(gray_conv, (7, 7), 0)
# get image dimensions:
gray_width = gray.shape[1]
gray_height = gray.shape[0]
focal_value = (gray_height / 2) / math.tan(math.radians(DFOV_DEGREES / 2))
# run detector:
result = cascade.detectMultiScale(gray)
for x, y, h, w in result:
dist = KNOWN_ROI_MM * focal_value / h
dist_in = dist / 25.4
# update display:
cv2.rectangle(image, (x, y), (x + w, y + h), (255, 0, 0), 2)
cv2.putText(image, 'Distance:' + str(round(dist_in)) + ' Inches',
(5, 100), cv2.FONT_HERSHEY_SIMPLEX, 1, (255, 255, 255), 2)
cv2.imshow('face detection', image)
if cv2.waitKey(1) == ord('q'):
break
run demo with:
python3 measure.py
-Jess


































